Gene regulation
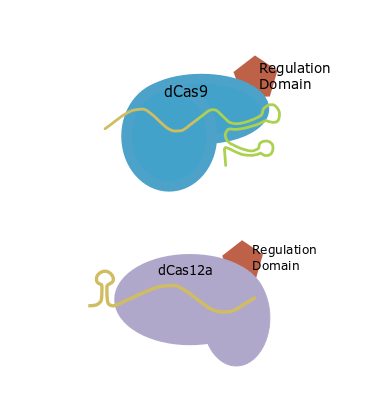
CRISPR tools can also be used as transcriptional regulators. GB has adapted the different strategies that can be used in transcriptional activation or transcriptional repression. To design your guides you must first choose one of the different dCRISPR strategies and then, the necessary and recommended parts will be shown. For transcriptional activation assays you can find a comparison of different strategies in Selma et al. (2019).
SAM
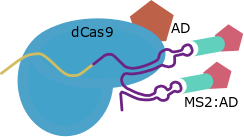
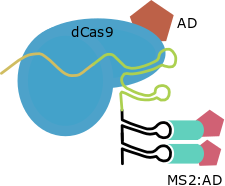
This strategy has been developed only for the structure of the Cas9 gRNA as a transcriptional activator. SAM use RNA aptamers added inside the loops of the gRNA scaffold as secondary anchoring sites and allows the design of Activation Domains combiantions due to this strategy maintains the direct fusion in C-Terminal. For this strategy other scaffold for the gRNA and the MS2 protein are required.
ScRNA2.0 and 2.1
This strategy has been developed only for the structure of the Cas9 gRNA as a transcriptional activator. scRNA use RNA aptamers added in the 3' of the gRNA scaffold as secondary anchoring sites and allows the design of Activation Domains combiantions due to this strategy maintains the direct fusion in C-Terminal. For this strategy other scaffold for the gRNA and the MS2 protein are required. The scRNA2.1 is an alternative version of the scRNA2.0 that shows high transcriptional activation levels (Selma et al,2019).
How to assemble single gRNAs for dCas9-based activation with GB?
1) Adapt your target sequence to the GB standard here. Dicot and monocot target sequences should start with 'G' and 'A' respectively. This constraint derives from the RNA polymerase III-dependent U6 and U3 promoters requirement of a 'G' or 'A' at the 5' end of the RNA to be efficiently transcribed. To avoid this constraint go to the polycistronic strategy to assemble a single gRNA preceded by a tRNA.
2) Assemble your GB-adapted target sequence with a PolIII promoter and the corresponding scaffold depending on the activation strategy you want to follow (see Table below) to create a guide RNA transcriptional unit here.
We advice you to use the scRNA2.1 scaffold since it gives the highest activation levels (Selma et al. 2019).
| Strategy |
Cas9 scaffold |
GB part |
| Direct fusion |
Basic scaffold |
GB0645 |
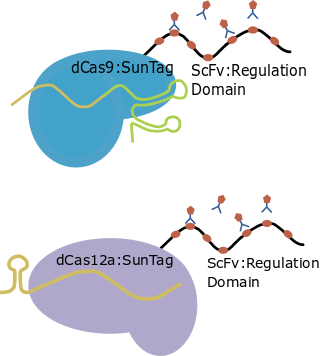
| SunTag |
Basic scaffold |
GB0645 |
| SAM |
MS2 aptamers inside Cas9 gRNA loops |
GB1436 |
| scRNA (PP7) |
PP7 aptamers in the 3' of the Cas9 gRNA scaffold |
GB1450 |
| scRNA (COM) |
COM aptamers in the 3' of the Cas9 gRNA scaffold |
GB1451 |
| scRNA2.0 (MS2) |
MS2 aptamers in the 3' of the Cas9 gRNA scaffold |
GB2461 |
| scRNA2.1 (MS2) |
MS2 aptamersin the 3' of the Cas9 gRNA scaffold (mutation on the aptamer that shows higher activation levels) |
GB1437 |
How to assemble polycistronic gRNAs for dCas9-based activation with GB?
Note: the polycistronic strategy is only available for the 'basic' and the 'scRNA2.1' scaffolds. We advice you to use the scRNA2.1 scaffold since it gives the highest activation levels (Selma et al. 2019) and therefore we will describe the assembly steps for this strategy.
1) Decide the number of gRNAs that you want to assemble in a single polycistron. With the scRNA2.1 scaffold either 1 or 3 gRNAs can be assembled in a single TU. These polycistronic RNA will be processed in planta by the endogenous tRNA processing system (RNase P and RNase Z) producing individual functional guide RNAs (tRNA-based strategy). See Xie et al. for details on the use of gRNA-tRNA arrays for multiplex genome editing.
2) Adapt your target sequences to the GB standard here. Number of target sequences to adapt to the GB standard will depend on the number of total gRNAs you want to include in the polycistron.
3) Clone in the pUPD2 a tRNA-protospacer-scaffold(2.1) unit for each position in the polycistron. For a 3X polycistronic RNA you will need three tRNA-protospacer-scaffold(2.1) units cloned in the pUPD2, each of them with a different part category (R1, Rn-1, RnC1). To do so click here.
4) Create a TU with a PolIII promoter and the previously assembled tRNA-protospacer-scaffold(2.1) units here.
5) Combine the TU created at 4) with extra single or polycistronic gRNAs and our recommended TUs/modules for gene regulation using the binary assembler.